How (do I plan) to get comfortable with Data Science as a Biologist
14 Jan 2018
I started with learning the programming language Python, and started with something easy:
- online courses available at DataCamp
- statistics book called ‘An Introduction to Statistical Learning, with Application in R’ by James, G., Witten, D., Hastie, T., and Tibshirani, R.
and of course talking with friends who uses Python.
‘Data Science’, ‘Machine Learning’, ‘Data Mining’, may seem like big confusing terms to a biologist. But once you read into it a little bit, we biologist are actually using ‘Data Science’ on a daily basis, albeit as what you can call a rather rudimentary form of it.
For example, when we want to do a Western blot, first we need to determine the protein concentration of each of our samples. We do that by finding a standard curve:
- have several protein samples with known concentrations ready
- use a color dye to stain proteins in these samples and the color intensity is linearly coorelated to protein concentration over a ‘useful range’ (for Western blot, in general it should be around 1 to 2 ug/ul)
- measure the color intensitiy
- plot protein concentration versus color intensity, and fit a line to the data i.e. do a Linear Regression
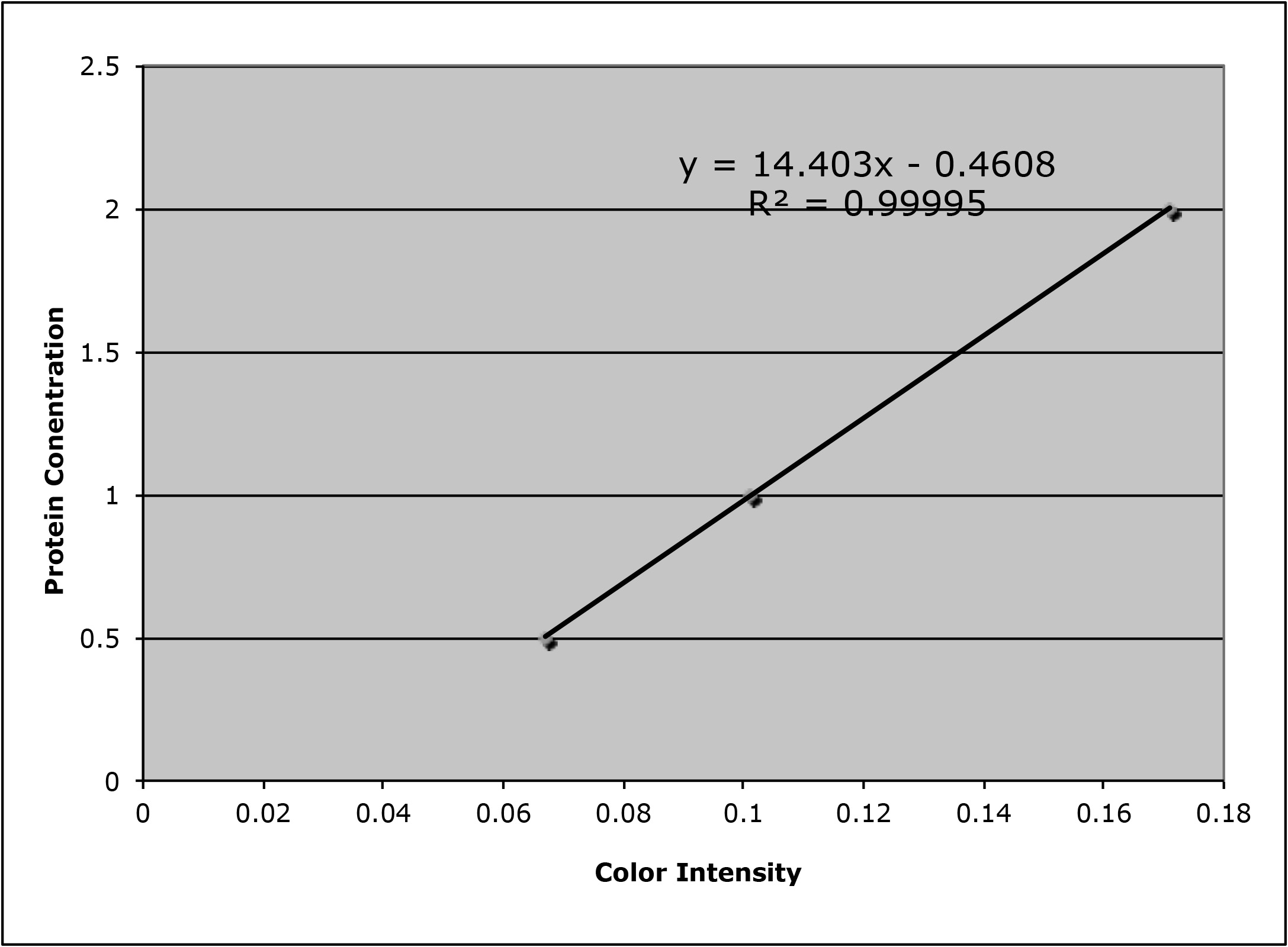
This is a very simplified version of ‘Machine Learning’ or alternatively called ‘Statistical Learning’ - which is to have a ‘training’ data set with known predictor values (color intensity) and response values (protein concentration). The standard curve (protein concentration versus color intensity) is a model that can then be used to predict the protein concentration of unknown samples!
The fun part, then, is learning about all those other mathematical and statistical models that can be use on all sorts of data. And when the predictor value is not just one, but tens of them, used to predict one outcome.